Novel insights into the structure of SARS-CoV-2 spike protein
– Sritama Bose
A salient feature of SARS-CoV-2, which causes COVID-19, is the presence of spike (S) proteins on the surface of the membrane that envelopes the genetic material of the virus. They appear as the characteristic crown-like spikes on the viral surface. The S protein is significant for multiple reasons: it mediates the entry of the virus into the host cell and is also the site where neutralising antibodies produced by the host cells bind to the virus in order to inactivate it. It is therefore important to understand the detailed structure of this protein.
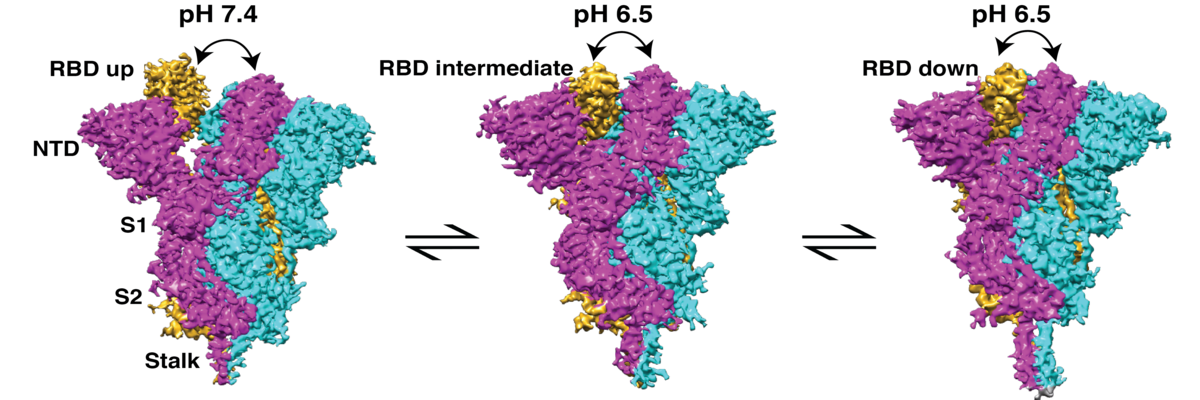
Most previous studies on the structure of the S protein have been carried out either at pH 8.0 or pH 4.0 to pH 5.0. But the structure of the S protein at physiologically relevant conditions ‒ at which the virus actually infects the host cells ‒ remains poorly understood. Recently, however, a research team led by Somnath Dutta, Assistant Professor at MBU, IISc, has made a crucial breakthrough. They have successfully visualised the different conformations or forms of the S protein at physiological pH 7.4 and near physiological pH (pH 6.5 and pH 8.0) using a technique called single-particle cryo-electron microscopy.
The S protein exists in mainly two conformations: open and closed. The open conformation is the form which helps the protein bind to the host cell receptor, thus making way for the virus to enter the cell. The team observed that around 68% of the S proteins exist in open conformation at physiological pH 7.4, but their proportion decreases when the pH is slightly higher (pH 8.0) or lower (pH 6.5). This suggests that the interaction between the S protein and receptor is more favoured at physiological pH (pH 7.4) than on either side of the biological pH scale.
In the study, the researchers also detected multiple distinct intermediate conformations between fully open and closed forms. Moreover, they show that distinct states of both conformations have different binding affinities towards neutralising antibodies.
The structural insights gained from this study could aid in developing therapeutics against SARS-CoV-2, including vaccines that target the S protein, according to the authors of the study.
REFERENCE:
Ishika Pramanick, Nayanika Sengupta, Suman Mishra, Suman Pandey, Nidhi Girish, Alakta Das, and Somnath Dutta, Conformational flexibility and structural variability of SARS-CoV2 S protein Structure, 2021, 29, 1-12.
https://doi.org/10.1016/j.str.2021.04.006
LAB WEBSITE:
https://somnath92.wixsite.com/website
Funding: Department of Biotechnology (DBT), Department of Science and Technology (DST-FIST)





