A tug-of-war between two enzymes drives spindle forces in cell division
Rohini Murugan
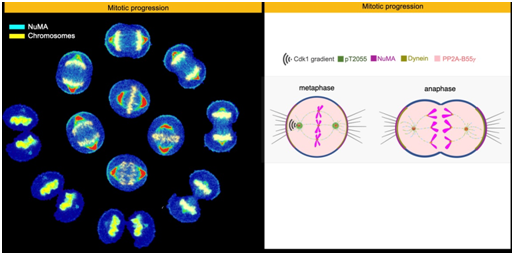
When eukaryotic cells divide, thread-like structures called spindle fibres help to pull a copy of the replicated chromosomes into each of the daughter cells. The forces responsible for this pulling should be generated at the right time to ensure error-free cell division. In animal cells, a protein that plays an integral role in this process is NuMA (Nuclear Mitotic Apparatus protein).
One of the ways in which cells regulate the localisation and functions of such proteins is by adding or removing phosphate groups. When NuMA is dephosphorylated at anamino acid residue called Threonine 2055 (T2055), it is localised to the cell cortex (part of the membrane) where it helps anchor the motor protein, dynein, which is essential for generating the spindle forces. However, enzymes like Cdk1 counteract this process by phosphorylating NuMA at this amino acid residue, to block its cortical localisation.
Very little is known about the dynamics of these two processes. Researchers from IISc have now identified and characterised the subunit (B55γ) of an enzyme called PP2A, which is responsible for dephosphorylation of NuMA at T2055. They have also identified other residues in the NuMA protein sequence that are critical for dephosphorylation.
The researchers suggest that a tug-of-war between the two enzymes, Cdk1 and PP2A-B55γ, regulates the cortical levels of NuMA. Since low levels of B55γ are linked to prostate cancer, future research will focus on unravelling the role of spindle formation in cancer progression.
Reference:
Riya Keshri, Ashwathi Rajeevan, Sachin Kotak. “PP2A-B55γ counteracts Cdk1 and regulates proper spindle orientation through the cortical dynein adaptor NuMA”, Journal of Cell Science (2020) 133: jcs243857.
doi: 10.1242/jcs.243857
Lab website: https://kotakcellbiology.wixsite.com/spindlebehaviour




