Assessing barriers in bacterial membranes
– Shatarupa Sarkar (with inputs from authors)
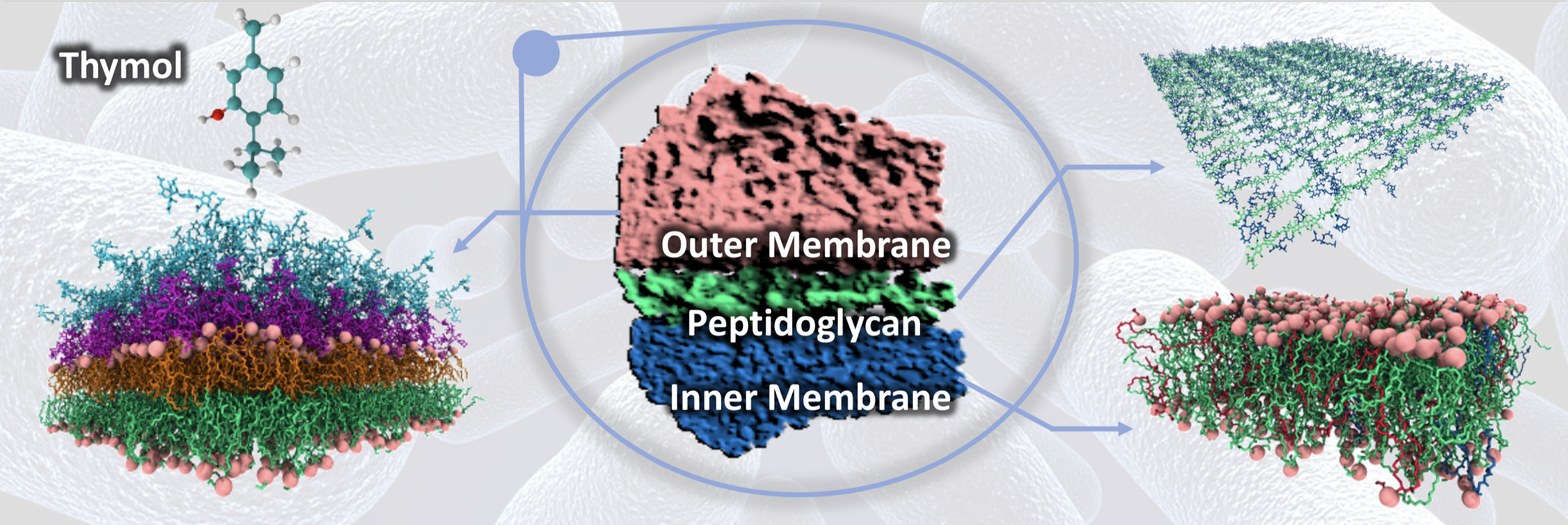
Bacteria are becoming increasingly resistant to available antibiotics; one of the reasons is their complex membrane, which has a multi-layered structure that has evolved to protect them from hostile environments. Some, like E. coli, have a more complicated membrane architecture than others and are composed of an inner phospholipid bilayer, an outer membrane consisting of lipopolysaccharides (LPS) and phospholipids with a peptidoglycan layer consisting of sugars and peptides sandwiched between the outer and the inner layers.
Researchers from the Department of Chemical Engineering and Department of Physics at IISc, in collaboration with Unilever Research and Development, have developed improved laboratory and computer simulation models to understand how antibacterial compounds can penetrate the bacterial membrane. Using the mobility of lipids as a marker for the entrance of thymol, an antibacterial molecule used in personal hygiene products, the group revealed the location of barriers present in the membrane, opening strategies to compare and improve the efficacy of antimicrobial formulations. They found that increased phospholipid content of the LPS layer allowed thymol to penetrate the cell membrane. These findings have been published by the American Chemical Society in the journal Langmuir.
The peptidoglycan layer provides mechanical strength to the bacterial cell and developing realistic computer models to study its properties for longer length and time scales using molecular simulations has been a grand challenge. A simplified model of the peptidoglycan layer developed by the simulations team is expected to reduce the computational effort by several hundred-fold, allowing one to study realistic models of the bacterial cell wall. Using this model, small molecules such as thymol were found to rapidly pass the peptidoglycan layer. This work has been published by the American Chemical Society in the Journal of Chemical Theory and Computation.
The study illustrates the ability to recreate laboratory and computational models that can be used to test the passage of small molecules through complex bacterial cell membranes. This opens up the possibility of screening potential antibiotics to assess their interaction with cell membranes, as well as the development of novel drug molecules to combat bacterial infections.
References:
Pradyumn Sharma, Srividhya Parthasarathi, Nivedita Patil, Morris Waskar, Janhavi S. Raut, Mrinalini Puranik, K. Ganapathy Ayappa, Jaydeep Kumar Basu. Assessing Barriers for Antimicrobial Penetration in Complex Asymmetric Bacterial Membranes: A Case Study with Thymol, Langmuir, 36, 30, 8800–8814, July 1, 2020.
Rakesh Vaiwala, Pradyumn Sharma, Mrinalini Puranik, and K. Ganapathy Ayappa.Developing a Coarse-Grained Model for Bacterial Cell Walls: Evaluating Mechanical Properties and Free Energy Barriers, J. Chem. Theory Comput.,16, 8, 5369–5384, July 6, 2020.
Lab websites:
https://kgalabiisc.wixsite.com/kgalab https://www.physics.iisc.ernet.in/~basu/




