How variability in sugar structures shields HIV
Human Immunodeficiency Virus (HIV) targets the immune system, specifically the CD4+ T cells, which are essential for fighting infections. If untreated, HIV can lead to Acquired Immunodeficiency Syndrome (AIDS). One of the biggest challenges in treating or preventing HIV infection is its high genetic variability.
HIV exists in multiple clades (or subtypes) that are spread across different continents worldwide, each with distinct genetic and structural features. The virus has a dense layer of glycans (sugar molecules) on its envelope protein (Env), which helps it evade the immune system by shielding key viral sites from antibody recognition.
A recent study from IISc addresses a crucial gap in HIV vaccine design: how glycan structures vary among HIV clades and affect the binding of broadly neutralising antibodies (bNAbs). The team used molecular dynamics simulations to model glycan behaviour on the Env protein across six major HIV clades.
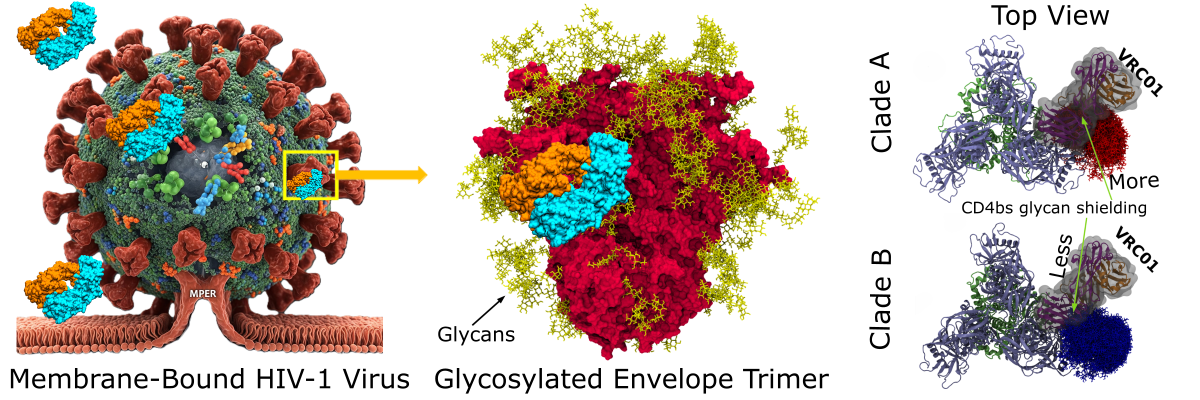
From left to right: HIV-1 virus (generated using GEMINI) bound to host-cell membrane. zoomed in view of spike protein (red) showing heavily glycosylated (yellow) envelope trimer bound with bNAb (cyan+orange) at CD4bs. VRC01 bNAb interaction with glycan at N462/N463; VRC01 (grey surface, purple: heavy chain, orange: light chain) bound to clade A and clade B Trimers. Clade A glycans at N462 are colored red (in the sticks), whereas clade B glycans at N463 are colored blue (in the sticks) (Image: Yogendra Kumar)
The study reveals that glycan shielding and accessibility to bNAbs vary significantly across clades. Even at conserved glycosylation sites, the number and flexibility of glycan conformers differ, affecting how bNAbs interact with the virus. bNAbs were found to interact with more glycans than previously recognised, and these interactions are clade- and antibody-class specific.
The study offers microscopic, quantitative insights into the dynamic and clade-specific role of the glycan shield. These findings are valuable for improving bNAb-based therapies and, more critically, for rational HIV-1 vaccine design strategies.
To elicit bNAbs with broad protection, vaccine immunogens must accurately mimic the Env protein and its glycan shield as it appears across different circulating clades. The observed clade-specific variations suggest that a single, one-size-fits-all immunogen may not be optimal for global coverage, highlighting the need for clade-optimised immunogens.
Cover Art Recognition by Journal:

Cover Art Caption:
The cover art illustrates escape of virus (green) from broadly neutralising antibodies (orange and red), facilitated by dense glycosylation (yellow) of the viral envelope glycoprotein (purple). This glycan shield impedes antibody binding, allowing infection and highlighting the structural complexity and adaptability of the virus in response to host immune pressure. The artwork was created using three different tools: Google’s Gemini AI, Visual Molecular Dynamics (VMD), and Inkscape. Google’s Gemini AI was used to generate specific elements of the cover art. The glycosylated HIV-1 Env trimer (colored yellow and purple) and the broadly neutralising antibody (bNAb) (colored orange and red) were visualised using VMD, utilising attributes such as MSMS for molecular surface representation. The glycans were rendered in Licorice style with a yellow color scheme, while the HIV-1 Env trimer was displayed in Surf style with purple coloring. Final adjustments and layout editing were completed using Inkscape (Image credit: Yogendra Kumar)
REFERENCE:
Arandhana M, Kumar Y, Dixit NM, Maiti PK, Clade-Specific influences of glycans on the interactions between HIV-1 envelope and broadly neutralizing antibodies, Computational Biochemistry (2025).
https://doi.org/10.1021/acs.jcim.5c01051
LAB WEBSITES:
https://physics.iisc.ac.in/~maiti/
https://chemeng.iisc.ac.in/telab/






